RNA sequencing and modification mapping by mass spectrometry
RNA modification is ubiquitous in extant life. Over 150 distinct post-transcriptional RNA modifications have been characterized. New approaches bringing together RNA sequencing, chemical manipulation, mass spectrometry and bioinformatics have expanded our understanding of the epitranscriptome and how it is affected by developmental and environmental changes. However, the precise location, stoichiometry, crosstalk, and function of most modifications remain elusive. The development of techniques for identifying the totality of ribonucleoside modifications and mapping their location is an area of active research with great potential for discovery and innovation. Current strategies are based on the specific cleavage of RNA using one or more endoribonucleases to generate suitable fragments for mass spectrometry analysis. Our research aims to improve experimental and computational protocols for unbiased RNA sequencing. In addition, we are eager to discover and characterize novel ribonuclease specificities and RNA modifying enzymes.
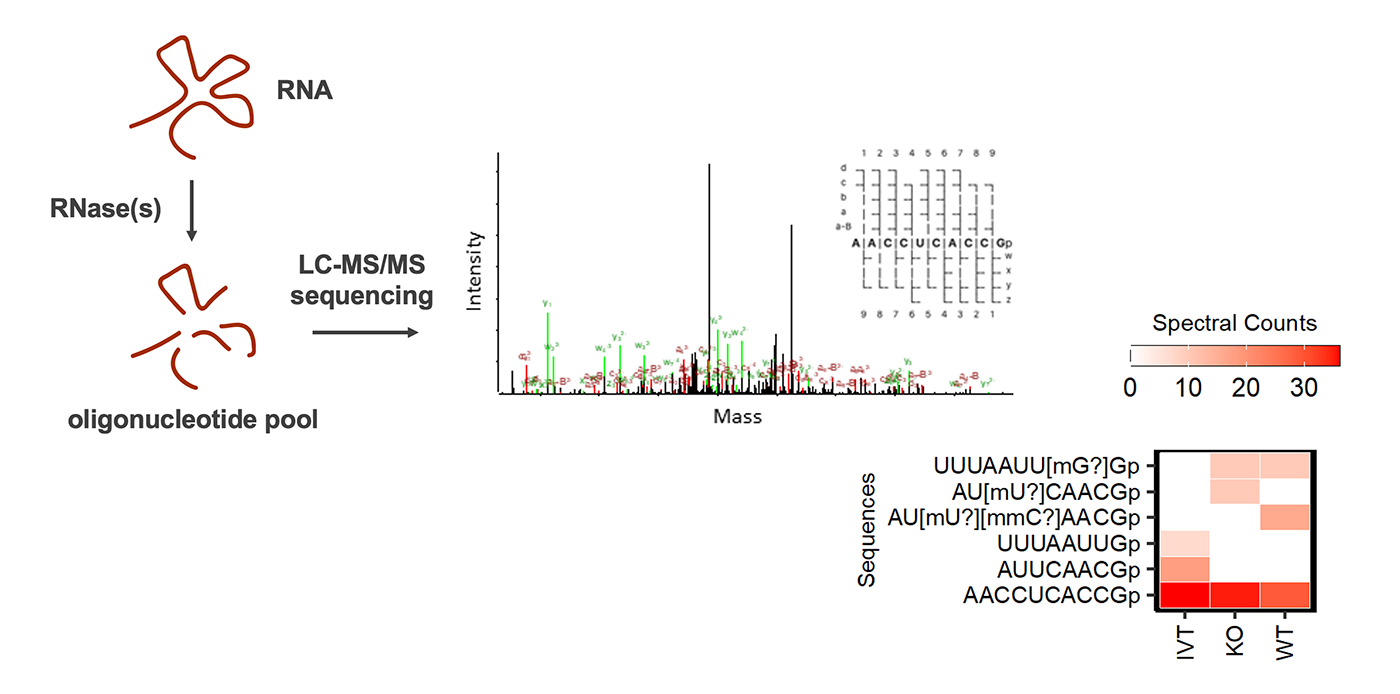
Schematic workflow for RNA modification mapping by mass spectrometry
We are actively pursuing innovative strategies for mapping RNA modifications through a combination of mass spectrometry and sequencing technologies. We apply comparative and de novo approaches to map modifications in synthetic and native RNA sequences. We are also advancing workflows to enable complete reconstruction of sequences and modification patterns in long, complex RNAs, including RNA-based therapeutics and vaccines.

