Making a Difference: Research on Neglected Tropical Diseases at NEB
Jeremy Foster, Ph.D., Sanjay Kumar, Ph.D., Larry McReynolds, Ph.D., Francine Perler, Ph.D., Barton Slatko, Ph.D., George Tzertzinis, Ph.D., Yinhua Zhang, Ph.D. and Clotilde Carlow, Ph.D.
NEB parasitology research group – the early days
In 1980, Dr. Comb recruited two scientists to conduct basic research projects at NEB – one researcher focused on restriction enzymes and the other worked on malaria. Five years later, a Parasitology Group was formed which accounted for approximately 50% of the research staff. The group’s focus soon shifted towards the poorly funded and understudied tropical diseases lymphatic filariasis and onchocerciasis. These diseases are caused by parasitic filarial nematodes and are transmitted by insect vectors. They are prevalent in over 80 countries, where over 150 million individuals are afflicted (Fig. 1). Currently, there are no vaccines for filarial diseases, but there are a few widely-used drugs (e.g., ivermectin, albendazole and diethylcarbamazine). However, these drugs only target larval stages and not the adult worms. Additionally, there are now signs of drug resistance developing to these commonly used drugs, causing great concern.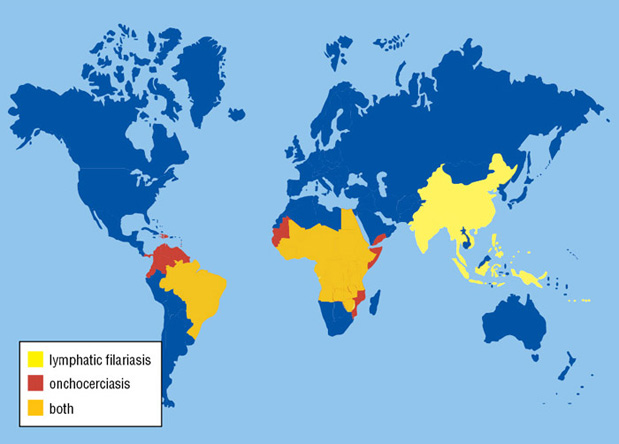
Overview of lymphatic filariasis and onchocerciasis
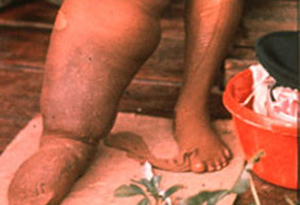
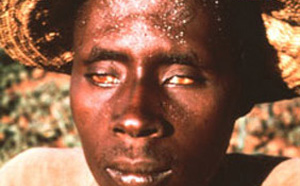
The World Health Organization (WHO) recognizes lymphatic filariasis as the third-leading cause of infectious disease-associated disability, after malaria and tuberculosis, while The Centers for Disease Control & Prevention (CDC) ranks onchocerciasis as the second-leading cause of infectious blindness after trachoma. These filarial diseases significantly impact the well-being of many individuals in the developing world, and contribute to the extreme poverty typical of endemic communities.In lymphatic filariasis, adult worms, such as Wuchereria bancrofti and Brugia malayi, mature and reproduce in the host lymphatic system, where they may remain for up to 10 years. Millions of larval worms are released into the bloodstream where they may be acquired by a female mosquito during a blood meal to initiate the insect-vector phase of parasite development. Infected individuals often present with kidney and lymphatic damage, where blockage of lymphatic ducts leads to the characteristic swelling (lymphedema) of limbs and genitalia. In severe chronic disease, gross disfigurement known as elephantiasis ensues (Fig. 2), causing disability, social stigma and psychological trauma. The related filarial nematode, Onchocerca volvulus, is the causative agent of onchocerciasis (river blindness) and is transmitted to humans by blackflies of the genus Simulium, which breed in fast flowing rivers. Long-lived adults reside and reproduce in host subcutaneous tissues, while the larvae released by female worms migrate to the skin and eyes, causing intense skin pathologies and eye lesions (Fig. 3).
Detection of filarial parasites: antibodies, antigens and DNA probes
NEB researchers in the Parasitology Group strive to apply the latest techniques to better understand the basic biology of filarial nematodes. In the early days, one such technique involved the construction of expression libraries for screening with various sera (1,2,3). Another approach involved radiolabelling parasite surface molecules to identify and characterize potentially protective antigens (4,5) and other molecules expressed at the host-parasite interface as candidate vaccine components and diagnostic tools. Vaccine studies were eventually carried out (6), and a protective nematode polyprotein allergen was identified (2). Immunoparasitological studies led to a method to identify mosquitoes transmitting B. malayi infection (7) and a biomarker for heartworm infection in animals (8,9). Molecular biology research resulted in the identification of a highly-repeated DNA sequence in B. malayi, which formed the basis of a genus-specific DNA probe to detect infection in humans (10,11).The genomics revolution: Wolbachia symbiont, drug targets and lateral gene transfer
The 1990s ushered in the era of modern molecular approaches to analyze parasite genomes. Fully sequencing metazoan parasite genomes of ~100 Mb was considered unrealistic at that time, so NEB joined the WHO/TDR-funded Filarial Genome Project, with a focus on parasite stage-specific gene discovery. cDNA libraries were constructed from different life cycle stages of B. malayi, and clones were partially sequenced to generate expressed sequence tags (ESTs). Through bioinformatic comparison to genes sequenced from other organisms, these ESTs shed light on biological processes in B. malayi (12). This was an early approach to transcriptome analysis that generated >25,000 ESTs from 25 cDNA libraries, and resulted in partial sequence information for nearly 10,000 clusters (genes) (13).During this analysis, bacterial sequences with similarity to Rickettsial bacteria were consistently observed and eventually attributed to the endosym-biotic bacterium Wolbachia, first suggested by EM studies in the 1960s, and confirmed by molecular means in 1995 (14,15). This maternally-inherited bacterium is an obligate mutualist in almost all filarial nematodes that infect humans, and has now been validated as a new drug target for filarial disease control in numerous human clinical trials using the antibiotic doxycycline (see (16) for review).
Anti-Wolbachia treatments cause a long lasting sterility of female worms but, more importantly, lead to adult worm death without any serious adverse effects in humans (16). However, due to difficulties of administering a protracted doxycycline treatment to large populations in endemic countries, alternative anti-Wolbachia therapies are being sought. To facilitate this approach, NEB scientists reported the full genome sequence of the Wolbachia endosymbiont from B. malayi in 2005 (17). A few years later, with advances in sequencing technology and throughput, members of the Parasitology Group also contributed to the full genome sequence of the nematode B. malayi (18). More recently, they published the first comprehensive identification of microRNAs (miRNA) in various developmental stages of B. malayi. This study forms the background for understanding miRNA function and the regulatory networks in the complex filarial life cycle (19). The availability of these and other genome sequences has enabled NEB researchers to develop rational drug target selection methodologies to generate a pipeline of potential drug targets (20,21). The rapid release of this data has provided scientists around the world with valuable resources that can highlight new approaches for attacking filarial parasites.
Several potential targets are being further investigated at NEB (22–30), and programs for new drug discovery are underway. We have incorporated forward and reverse genetic approaches using the genetically tractable free-living nematode Caenorhabditis elegans. This has enabled the validation of selected nematode targets and experimental characterization that would be impossible in parasitic species (26,31,32,33).
Comparison of the B. malayi and Wolbachia genome sequences identified metabolites that might be sourced from one organism to the other and thereby underlie the mutualistic symbiosis. Among these are nucleotides, riboflavin and heme. B. malayi is deficient in de novo synthesis of these molecules, while Wolbachia has retained biosynthetic capabilities despite having a streamlined genome typical of most endosymbionts (17,18).
A role for Wolbachia in meeting its nematode host’s requirement for riboflavin and heme has been confirmed through experimentation at NEB (29,34), validating the biosynthetic pathways leading to these metabolites as potential drug targets. The riboflavin pathway is fully functional in Wolbachia, and implicated a nutritional role for the endosymbiont by showing supplementation with this vitamin partially rescued filarial parasites treated with doxycycline to remove their Wolbachia. With respect to the heme biosynthetic pathway, B. malayi contains only ferrochelatase, the enzyme that catalyzes the last step in the pathway. We discovered that the gene encoding ferrochelatase is the result of lateral gene transfer (LGT) from a Rhizobiale bacterium. The protein product is both functional and essential for B. malayi (31). Further genome comparisons have revealed a surprisingly high level of trans-kingdom LGT, with about 15% of the Wolbachia genome also being present in the B. malayi genome (35,36). The LGTs include over 30 full-length genes, some of which appear to be transcribed at high levels in a life-cycle stage-specific manner.
New approaches, new technologies
Ongoing areas of parasitology research at NEB are exploiting the post-genomics revolution and associated emerging technologies to address new questions and re-visit old problems with contemporary approaches. For example, an oligo capture method has been adapted to enable targeted sequencing of additional Wolbachia genomes, without the need to simultaneously sequence the 100-fold larger nematode host genome (37). This allows quick and efficient sequencing of Wolbachia from different clinical isolates and from other filarial nematodes.Various other ‘omics studies are being pursued at NEB. These include transcriptomics (RNA-seq) and microRNA analysis (19) of multiple life cycle stages of different filarial nematodes and their Wolbachia endobacteria, as well as tissue-specific transcriptomics. Proteomic studies are also being conducted with a focus on the nematode surface glycoproteome – a research area that featured in the early days of parasitology at NEB, as well as the feeding apparatus of mosquito vectors.
Filarial diagnostics also featured prominently some 30 years ago, and is now benefiting from state of the art, modern DNA-detection technologies. Loop-mediated isothermal amplification (LAMP) of filarial-specific sequence results in a visual read-out of amplification with high sensitivity and specificity without the need for sophisticated instrumentation (38), making the approach well-suited to field use in endemic countries.
Filarial nematodes have previously been intractable to genetic manipulation, but recent advances in this area by NEB scientists are opening the door to studies not possible a few years ago. These include a more efficient method for silencing genes by RNA interference (RNAi) (39). This involves soaking worms in a heterogeneous mix of short interfering RNAs, where the molecules in the mixture are produced enzymatically from a longer dsRNA with the correct size and end modifications for efficient silencing (40,41). Methods for whole-mount immunofluorescence have also been developed for examining, in detail, the cell biology of Wolbachia and filarial nematodes in both wild-type and RNAi-treated worms (39,42). A system for transfecting B. malayi larvae has also been developed, in which the transfected parasites maintain transgene expression around the life cycle and into the subsequent generation (43). It is hoped that these studies will pave the way for performing future genetic analyses directly in filarial nematodes.
Perspective
For nearly 35 years, parasitologists at NEB have applied the latest biochemical and molecular techniques to the study of malaria, filarial nematodes and their Wolbachia endosymbionts in the areas of genomics, vaccines, diagnostics and drug targeting. The aim throughout has been to uncover and freely disseminate details concerning the basic biology of these organisms to help the research community achieve the goal of eliminating these neglected parasitic diseases.References
- Arasu, P., et al. (1987) Exp Parasitol. 64, 281–291.
- Poole, C.B., et al. (1992) Proc Natl. Acad. Sci. U.S.A. 89, 5986–5990.
- Tuan, R.S., et al. (1991) Mol. Biochem. Parasitol. 49, 191–203.
- Edwards, M.K., et al. (1990) Trop. Med. Parasitol. 41, 174–180.
- Storey, N, and Philipp, M. (1992) Exp. Parasitol. 74, 57–68.
- Carlow, C.K, and Philipp, M. (1987) Am. J Trop. Med. Hyg. 37, 597–604.
- Carlow, C.K., et al. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6914–6918.
- Hong, X.Q., et al. (1996) Parasitology. 112, 331–338.
- Santiago Mejia, J., et al. (1994) Parasite Immunol. 16, 297–303.
- McReynolds, L.A., et al. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 797–801.
- Williams, S.A., et al. (1993) Exp. Parasitol. 77, 235–245.
- Williams, S.A., et al. (2000) Int. J. Parasitol. 30, 411–419.
- Elsworth, B., et al. (2011) Int. J. Parasitol. 41, 881–894.
- Kozek, W.J. and Marroquin, H.F. (1977) Am. J. Trop. Med. Hyg. 26, 663–678.
- Sironi, M., et al. (1995) Mol. Biochem. Parasitol. 74, 223–227.
- Foster, J.M., et al. (2013) The Wolbachia Bacterial Endosymbionts of Filarial Nematodes. In: Kennedy MW, Harnett W, editors. Parasitic Nematodes Molecular Biology, Biochemistry and Immunology. 2nd ed. Wallingford: CABI. pp. 308-336.
- Foster, J., et al. (2005) PLoS Biol. 3, e121.
- Ghedin, E., et al. (2007) Science, 317, 1756–1760.
- Poole, C.B., et al. (2014) PLoS One, 9, e96498.
- Holman, A.G., et al. (2009) BMC Microbiol. 9, 243.
- Kumar, S., et al. (2007) PLoS ONE, 2, e1189.
- Farelli, J.D., et al. (2014) PLoS Pathog. 10: e1004245.
- Foster, J.M., et al. (2009) Parasitol. Res. 104, 1047–1052.
- Gloeckner, C., et al. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3424–3429.
- Li, Z., et al. (2011) PLoS Negl. Trop. Dis. 5, e1411.
- Novelli, J.F., et al. (2009) Dev. Biol. 335, 340–355.
- Raverdy, S., et al. (2008) Mol. Biochem. Parasitol. 160, 163–166.
- Raverdy, S., et al. (2007) Mol. Biochem. Parasitol. 156, 210–216.
- Wu, B., et al. (2009) PLoS Negl. Trop. Dis. 3, e475.
- Tzertzini, G., et al. (2010) PLoS Negl. Trop. Dis. 4, e2625.
- Wu, B., et al. (2013) Proc. Natl. Acad. Sci. U.S.A. 110, 7748–7753.
- Zhang, Y., et al. (2004) J. Biol. Chem. 279, 37185–37190.
- Zhang, Y., et al. (2005) Dev. Biol. 285, 330–339.
- Li, Z., and Carlow, C.K. (2012) PLoS One, 7, e51597.
- Hotopp, J.C., et al. (2007) Science, 317, 1753–1756.
- Ioannidis, P., et al. (2013) BMC Genomics, 14, 639.
- Geniez, S., et al. (2012) Symbiosis, 58, 201–207.
- Poole, C.B., et al. (2012) PLoS Negl. Trop. Dis. 6, e1948.
- Landmann, F., et al. (2012) Parasit. Vectors. 5, 16.
- Morlighem, J.E., et al. (2007) Biotechniques, 42, 599–600.
- Xiao, J., et al. (2009) RNA, 15, 984–991.
- Xiao, J., et al. (2009) RNA, 15, 984–991.
- Landmann, F., et al. (2010) PLoS Negl Trop Dis 4, e758.
- Xu, S., et al. (2011) Int. J. Parasitol. 41, 355–362.

